Posted on February 27, 2011
Species of fungi can usually be distinguished based on what they look like. This method is known as the morphological species concept (study of the form, structure and configuration of organisms). But not all fungi can be studied in this way.
Some fungi that occupy the same functions in ecosystems look identical, even though they are different species. Such species are cryptic, because they cannot be distinguished from each other with morphology. One way of dealing with such species is to apply the biological species concept, which states that if two individuals can mate with each other and produce fertile offspring, they should be the same species. This concept is useful if the sexes exist in different individuals, but some fungi are self-fertile – an individual can mate with itself or with another individual, and it is very difficult to distinguish these two events in nature.
One such fungal organism is known as Chrysoporthe austroafricana (the Latin for “orange destroyer from southern Africa”). This fungus is closely related to Chrysoporthe cubensis (orange destroyer from Cuba, although it occurs throughout South America). Both can self and both of them cause a serious stem disease of plantation Eucalyptus (blue gum) trees, which can weaken the stem and cause the tree to die or fall down during high wind. Chrysoporthe cubensis is also present in Southeast Asia, where it causes an identical disease. Scientists thought for a long time that the fungus from South America and the one from Southeast Asia might be different species, simply because of the vast distances between them, but evidence for this separation was not forthcoming.
 |
|
| An example of the devastation that Chrysoporthe austroafricana can cause in gum tree plantations in South Africa. This plantation of susceptible Eucalyptus clones are approximately 40% infected with the fungus, and some trees such as the one in the center have already fallen down after high wind conditions. Albe van der Merwe (right) is busy sampling C. austroafricana from the base of a dead young tree, and he is assisted by Daleen van Dyk. |
Top: Sexual fruiting structures of Chrysoporthe cubensis from Colombia, with spore drops oozing from the necks of perithecia. Bottom: Albe van der Merwe beside a 30-year-old Eucalyptus grandis tree with a stem infection by Chrysoporthe austroafricana clearly visible. Although such old trees are not easily toppled by wind, the fungus girdles the stem and causes the tree to die. |
In a paper by Van der Merwe and coworkers[i] (Department of Genetics and Forestry and Agricultural Biotechnology Institute, University of Pretoria), this problem was finally resolved. They used population genetics to show that populations of C. cubensis from South America and Southeast Asia are genetically different, and do not exchange genetic material. Armed with this knowledge, they searched for a quicker and easier way of distinguishing the species, because it is impractical to perform population genetic analyses in order to identify a single individual.
By sequencing the beta-tubulin gene, which is a gene that codes for proteins that form part of the cytoskeleton of a cell, two signature DNA sequences were uncovered that could aid to differentiate the three species of Chrysoporthe. When specific enzymes are added to amplifications of this gene from individuals of the fungi, they differentially cut the DNA at these sites, forming different sizes of fragments. These fragments could then be separated based on size, and a profile was obtained. This represented a breakthrough in developing a quick identification technique, because most laboratories have the equipment and expertise needed to perform the technique.
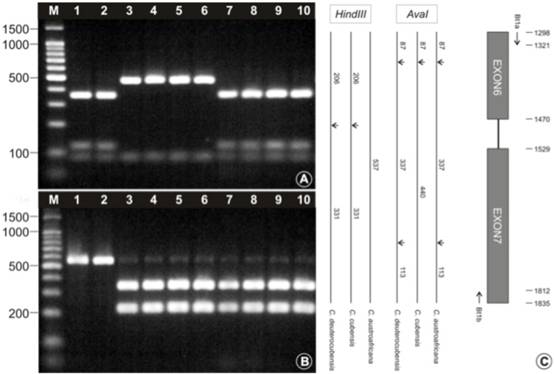 |
| A “map” of the beta-tubulin gene of C. austroafricana, C. cubensis and C. deuterocubensis can aid to distinguish these species quickly and easily. A and B show the profiles obtained using two different restriction enzymes (HindIII and AvaI). 1-2, Chrysoporthe austroafricana; 3-6, Chrysoporthe cubensis; 7-10, Chrysoporthe deuterocubensis. C shows a map of the beta-tubulin gene region, the sites where the enzymes cut the DNA, and the sizes of the DNA fragments that are produced. |
Now that the species could be distinguished from each other, the final task was to describe the fungus from Southeast Asia as a new species. But there was a problem, because it looks exactly the same as C. cubensis from South America and causes the same disease, and it can self fertilize. It was thus impossible to apply either the morphological or the biological species concept. Fortunately, scientists have recently started to use DNA identification techniques to describe cryptic species in other fungi, although only a few such fungi have been described with new names. The UP scientists followed this trend and used information from the quick identification technique to convince the scientific community that these three species really are different. The fungus was thus described as Chrysoporthe deuterocubensis (the “other” cubensis).
The new information gathered by UP scientists has important implications for quarantine and the continued prosperity of forestry in South Africa. The unholy trinity of gum tree pathogens is represented by three species of Chrysoporthe, each of which occurs in a different part of the world. Trade in wood and wood products could introduce one of these species to another part of the world, where it could intensify disease on forest trees. In fact, both C. cubensis and C. deuterocubensis have already been introduced onto the African continent, although those populations are far north of South Africa. Nobody knows what the effect will be when these fungi reach South Africa, and that is why it is important to never transport wood between different countries without a permit.|
[i] Van der Merwe, N.A., E.T. Steenkamp, M. Gryzenhout, B.D. Wingfield and M.J. Wingfield. 2010. Multigene phylogenetic and population genetic data confirm the existence of a cryptic species in Chrysoporthe cubensis. Fungal Biology 114: 966-979. |
|
Copyright © University of Pretoria 2024. All rights reserved.
Get Social With Us
Download the UP Mobile App