We employ several strategies including elucidation of drug target structure-activity properties of proteins involved in underexplored metabolic pathways. Additionally, genome-wide investigation of the transcriptome, proteome and metabolome of the parasite after perturbation of these metabolic pathways is employed, allowing explorations of the gene regulatory mechanisms the parasite utilize to control its asexual proliferation in human erythrocytes.

In a project funded by the South African Malaria Initiative, we focus on essential and /or unique proteins of the malaria parasite that have been selected as potential drug targets. Both atomic resolution structural analyses and in silico predictors are used to guide structure-based descriptions of novel inhibitors for these drug targets, which are then tested in vitro. This project is collaboratively run with Braam Louw (Emeritus Prof associated with the group) and Steven Pelly (University of Stellenbosch).


Additionally, we explore previously unknown biological phenomena including investigating polyamine transport mechanisms as possible drug targets or drug delivery mechanisms. Collaboration with Prof K Kirk (Australian National University, Australia), Prof Otto Phanstiel (University of South Florida) and Animex Corp, USA.
We are currently focussed on the antimalarial activity of biogenic amines, which form a novel structural class of compounds not previously shown to have antimalarial activity. The compounds are active against other parasites and cancerous cells as well, and seem to elicit their efficacy through targeting essential epigenetic control mechanisms of cell cycle development. In the malaria parasite, these compounds are highly active (20-70 nM) and selectively only kills the parasite and not mammalial cells (>7000 selectivity). The compounds undergo iterative medicinal chemistry guided optimisations for optimal in vivo efficacy. Work supported by the South African Malaria Initiative in collaboration with Pat Woster and Kip Guy (Medical University of South Carolina and St Judes Children’s Hospital, USA) and Lubbe Wiesner (University of Cape Town).

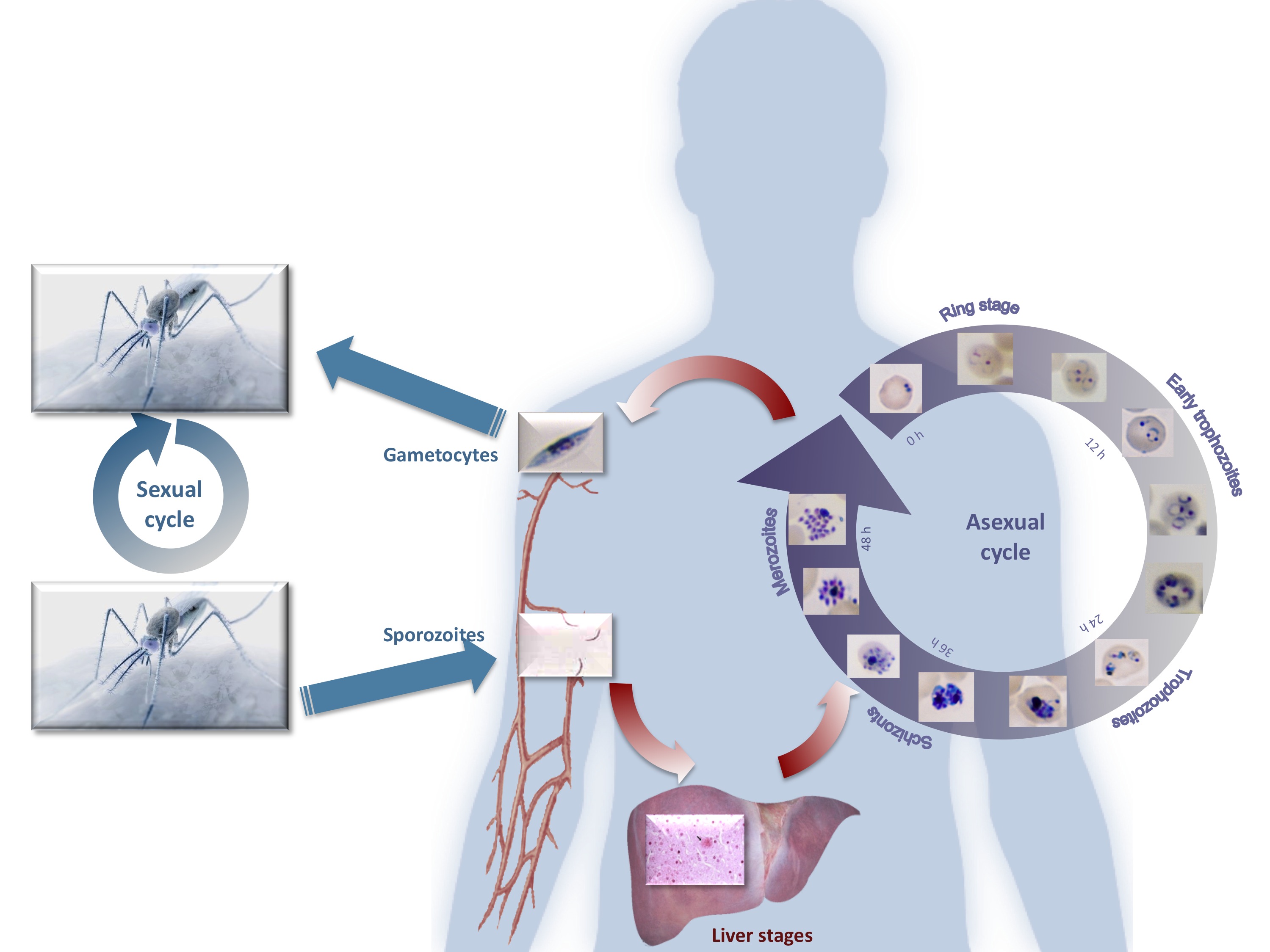
Genomic mapping of cell cycle regulators of malaria parasites:
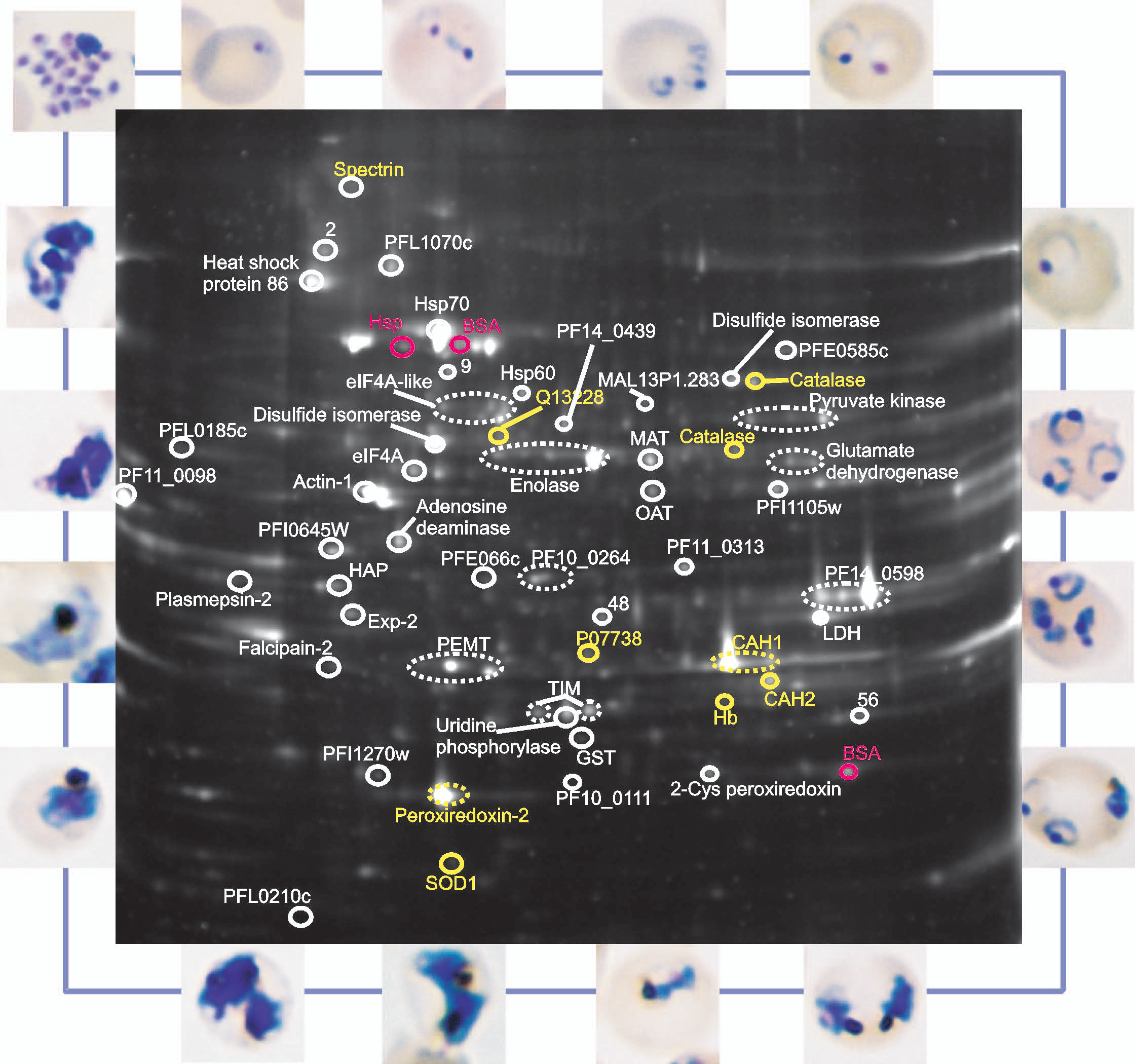
Malaria parasites undergo massive replication to billions of parasites in human red blood cells, resulting in the characteristic flu-like pathogenesis of the disease. In a project funded by the EU (FP7 Network of Excellence, Evimalar), genomic signatures and signalling pathways contributing to cell cycle control during this replication cycle of malaria parasites are under investigation. Transcriptome analysis (RNAseq) of parasites halted in their cell cycle progression reveal target control points that could exploited for efficient blocking of parasite proliferation, and therefore reveal novel antimalarial drug targets.
Enabling transmission blocking to eliminate malaria:
Determination of mode-of-action of antimalarials against transmission blocking forms of the parasite:
In a project supported by the South African Malaria Initiative, we are establishing assays to predict the mode-of-action of new compounds for which activity has been shown against gametocytes. This includes determining viability, rate of killing, stage-specificity and biochemical pathways that are targeted. The work makes part of a Gauteng transmission blocking platform in collaboration between UP, CSIR, and WITS.
Specific forms of the parasite develop in humans and these gametocytes are transmitted to mosquitoes to continue the spread of the disease. In order to eliminate malaria globally, these transmittable forms of malaria need to be targeted. In a collaborative effort (UP, CSIR and WITS), the Gauteng Transmission Blocking Platform investigates novel drugs targeting malaria gametocytes, funded by the South African Malaria Initiative and the Medicines for Malaria Venture. As part of understanding the biology of gametocytes, genomic profiling of drug sensitive and resistant parasites is investigated, including parasites isolated from malaria infective patients.
Copyright © University of Pretoria 2024. All rights reserved.
Get Social With Us
Download the UP Mobile App